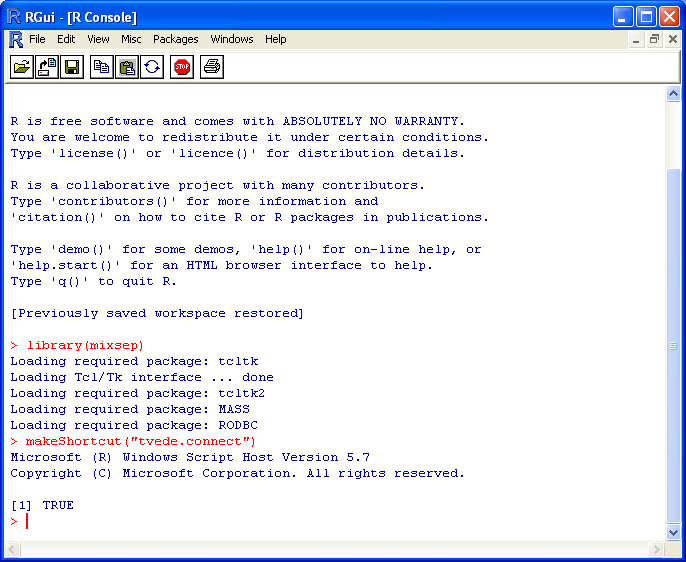
In order to do so one needs to give a connection file as argument to the makeShortcut-function (see the shortcut guide). One example of such a file is given below (the file name and extension is unimportant). The mixsep GUI uses the odbcDriverConnect-function from the RODBC-package in order to connect to a database. Hence, please refer to the documentation for this function by typing ?odbcDriverConnect in R or read the package Vignette by Brian Ripley.
The content of connection file (tvede.connect). It is important that only one argument (db, dbtab, dbcase, dbcols) is specified per line. Each argument is given in double-quotes ("argument") and these are separated by semi-colon ("argument1; argument2"). Each line is described in more detail below:
The screen short below show the creation of a "mixsep" desktop shortcut
while creating the database connection.
NOTE:
The connection file (here tvede.connect)
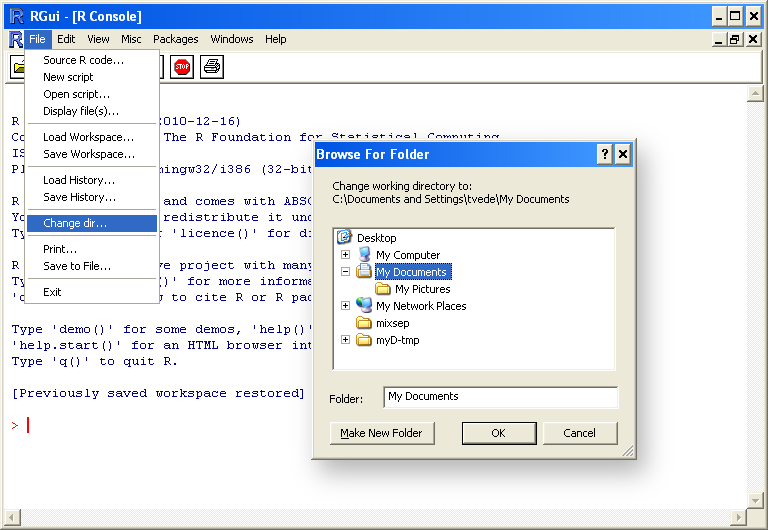
needs to be in the current working directory, which is set by
selecting "Change dir..." under "File" in the menu (se below). The
chosen directory should contain the connection file.

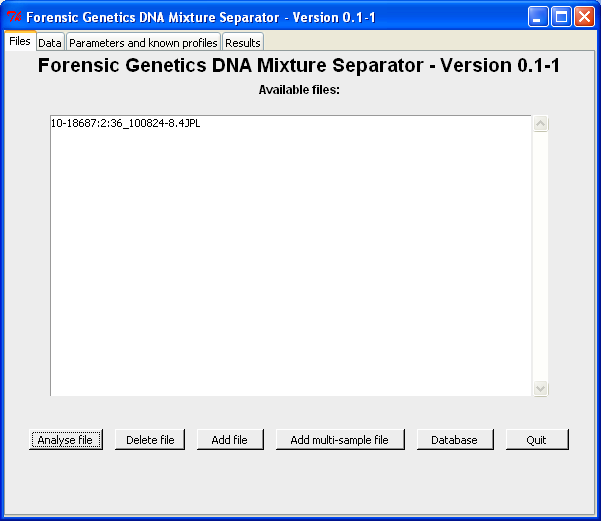
By clicking this button a "database query"-window opens in which queries to the specified database can be made:
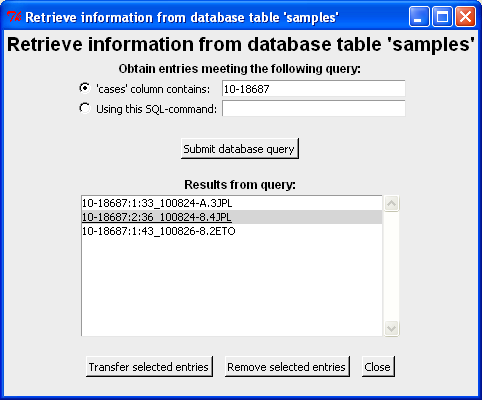
In the screen shot below a query to the case 10-18687 is made and three different analysis results are returned. Furthermore, the call to the database uses wild cards such that any consecutive substring of 10-18687 would yield the three samples (and possibly more). Note that ":" is used to separate the column information from the columns specified in dbcase and dbcols above. Each row corresponds to an unique sample file (originating from different replicates and PCR runs).
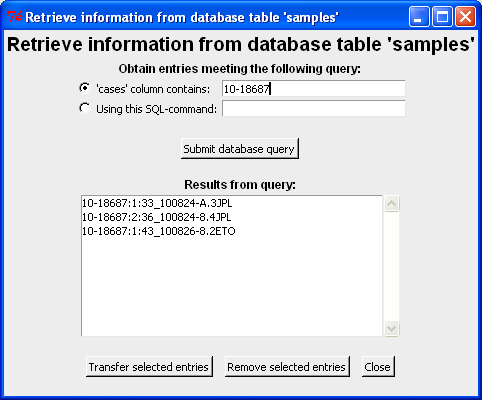
The sample(s) of interest can be transferred to the main window by double-click a the row or by highlighting the row(s) and click "Transfer selected entries". Note both [Shift], [Ctrl] and [Ctrl-a] can be used to select more/all samples. Here we select 10-18687:2:36_100824-8.4JPL for further analysis (which is then transferred to the main window - see below):